Search CLASH, Gene Expression, or microRNA Expression
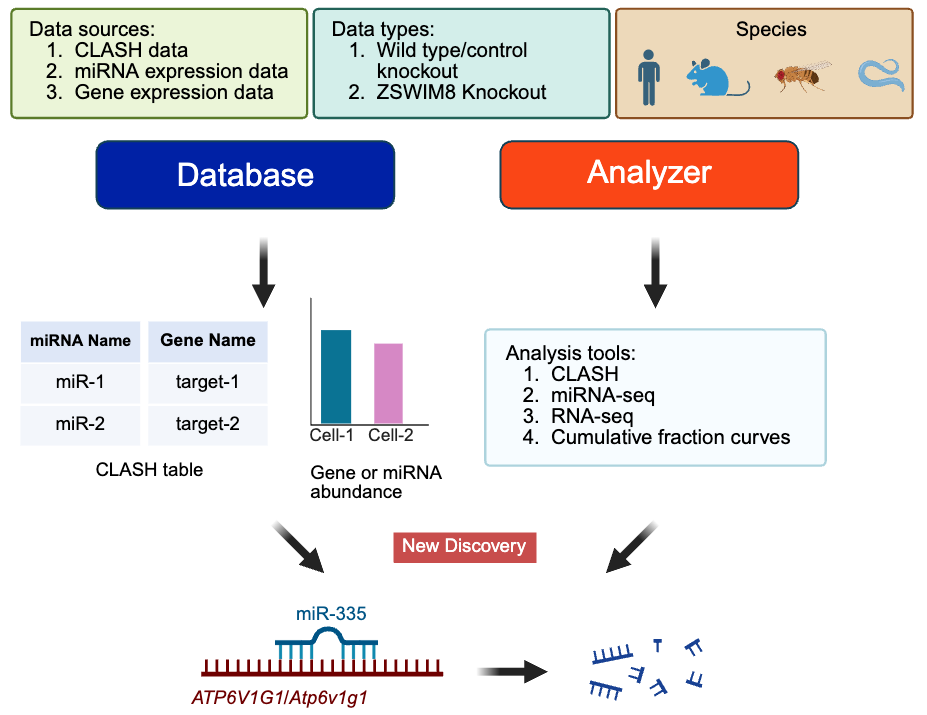


CLASHub 1.0 is a comprehensive and user-friendly platform for exploring microRNA-target interactions. It hosts CLASH (Crosslinking, Ligation, and Sequencing of Hybrids—a method to directly identify miRNA-target interactions bound to AGO proteins) data from 27 distinct cell lines or tissues across four model organisms (Human, Mouse, Drosophila, and C. elegans), comprising over 170 samples. The database includes 80+ published CLASH samples and 90+ newly generated datasets, including 15 cell lines or tissues not previously reported. Among them are 50 ZSWIM8 knockout samples—providing valuable insights into target-directed microRNA degradation (TDMD).
CLASHub also offers extensive expression profiling: 260 RNA-seq samples from 51 cell lines or tissues, and 168 miRNA-seq samples across 25 cell types. These datasets enable integrated analyses of miRNA-target regulation across diverse biological contexts.
Beyond serving as a database, CLASHub features a powerful Analyzer tool for simple, fast, and in-depth analyses—including CLASH data processing, miRNA-seq analysis, RNA-seq analysis, and cumulative fraction curve generation. These tools support precise identification of miRNA binding sites and quantitative analysis of miRNA impact on gene expression.
Using CLASHub, we identified ATP6V1G1 as a novel trigger for miR-335 degradation, a TDMD-sensitive miRNA, validated in both human and mouse cells.
By integrating high-quality data with robust analytical tools, CLASHub facilitates comparative studies across multiple organisms, advancing the understanding of miRNA-mediated gene regulation in health and disease.
If you use CLASHub in your research, please cite:
Li L, Sheng P, Hiers NM, Li T, Grimme AL, Wang Y, Traugot CM, Xie M. CLASHub: an integrated database and analytical platform for microRNA-target interactions. bioRxiv, 2025. https://doi.org/10.1101/2025.08.05.668543